Operons:
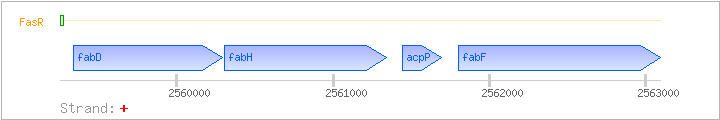
fabDHPF
| fabD |
SCO2387 |
SC4A7.15, fabD |
+ |
2559340..2560290 |
malonyl CoA:acyl carrier protein malonyltransferase |
| fabH |
SCO2388 |
SC4A7.16, fabH |
+ |
2560307..2561338 |
3-oxoacyl-[acyl-carrier-protein] synthase III |
| acpP |
SCO2389 |
SC4A7.17, acpP |
+ |
2561443..2561691 |
acyl carrier protein |
| fabF |
SCO2390 |
SC4A7.18, fabF |
+ |
2561801..2563090 |
3-oxoacyl-[acyl-carrier-protein] synthase II |
| Operon evidence: |
RT-PCR, S1, Disruption
|
| Reference: |
Revill WP, et al. (2001)
|
| Comments: |
In this study we provide further biochemical evidence
for the role of a small cluster of presumed fab genes (which
includes fabH) in Streptomyces coelicolor and a transcriptional analysis of the fab cluster, and we have attempted to disrupt fabH to determine if it is essential for the viability of the cells.
|
Promoters
| FasR |
- |
Positive |
-87:-67(from translation start site) |
2559253..2559273 |
TTTTGTGCAGGGTCCACAAAA |
Xu D, et al. (2010): OV, HM, GS, qRT-PCR
|
| Comments:
|
| FasR |
A novel transcription factor, FasR (SCO2386), controls
expression of fabDHPF operon and lies immediately
upstream of fabD, in a cluster of genes that is highly
conserved within actinomycetes. Expression of fab
genes was downregulated in the fasR mutant, indicating
a role for this transcription factor as an activator. FasR binds specifically to a DNA sequence containing fabDHPF promoter region, both in vivo and in vitro. |
Terminator
No Data
Overview
|
|
