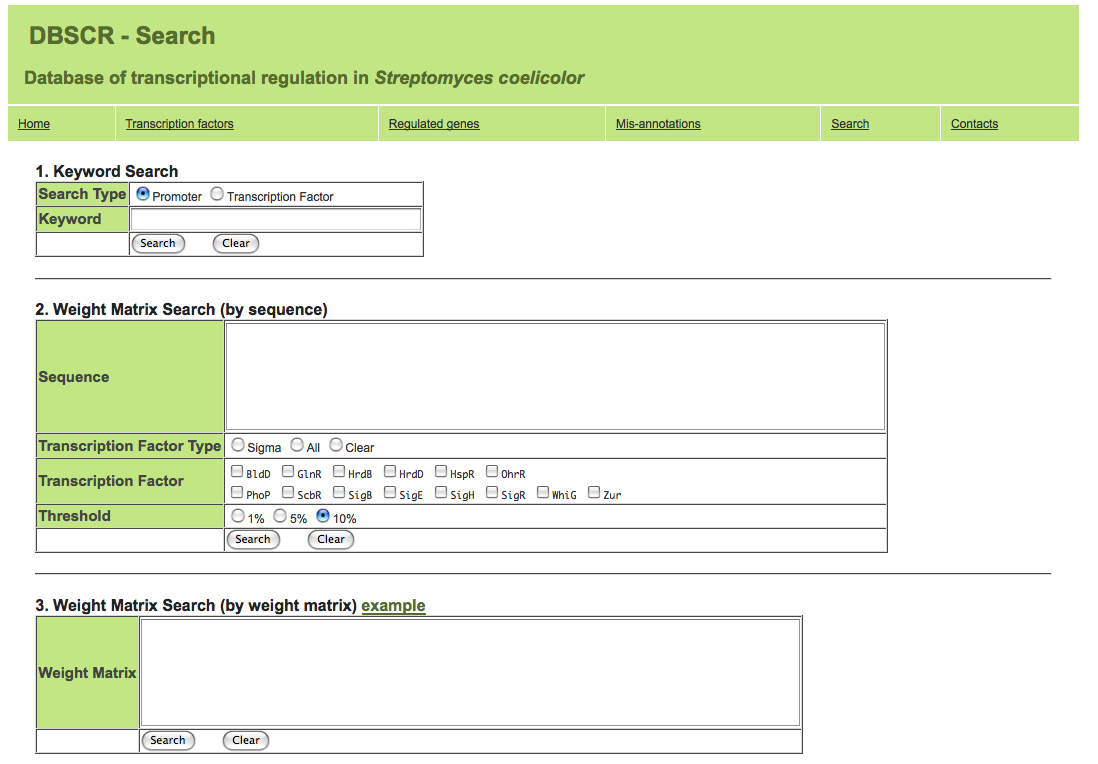
Search function
1. Keyword Search
Search for transcription factor or regulated genes with gene name (ex. ahpC, SCO5032).
2. Weight Matrix Search (by sequence)
This search function may help to find transcription factor binding DNA motif in your input promoter sequence. Please enter promoter seqrucne with fasta format or only sequence.
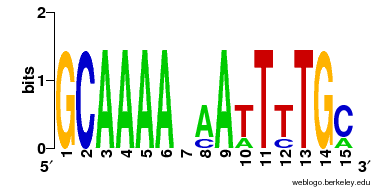
3. Weight Matrix Search (by weight matrix)
This program returns the top 10 similar weight matrices against your input weight matrix.
Use a tab (or space) deliminated format.
The first line, if starting with ">", is ignored.
Both absolute numbers and relative frequencies are acceptable.
Input nucleotides should be in the order A->C->G->T.
|
|
Example format 1:
>RocR
0 0 7 7 7 7 1 6 7 2 0 0 0 0 1
0 7 0 0 0 0 0 1 0 0 0 1 0 0 6
7 0 0 0 0 0 2 0 0 0 0 0 0 7 0
0 0 0 0 0 0 4 0 0 5 7 6 7 0 0
Example format 2:
0 0 1 1 1 1 0.143 0.857 1 0.286 0 0 0 0 0.143
0 1 0 0 0 0 0 0.143 0 0 0 0.143 0 0 0.857
1 0 0 0 0 0 0.286 0 0 0 0 0 0 1 0
0 0 0 0 0 0 0.571 0 0 0.714 1 0.857 1 0 0