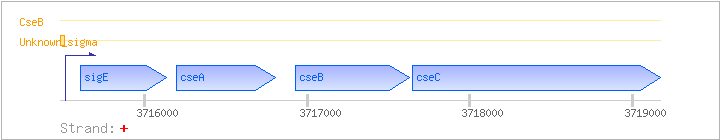
Operons:
sigE
sigE-cseABC
| sigE |
SCO3356 |
SCE94.07, sig28, sigE |
+ |
3715604..3716137 |
ECF sigma factor |
| sigE |
SCO3356 |
SCE94.07, sig28, sigE |
+ |
3715604..3716137 |
ECF sigma factor |
| cseA |
SCO3357 |
SCE94.08, cseA |
+ |
3716194..3716802 |
hypothetical protein |
| cseB |
SCO3358 |
SCE94.09, cseB |
+ |
3716926..3717630 |
two-component system response regulator |
| cseC |
SCO3359 |
SCE94.10, cseC |
+ |
3717647..3719170 |
putative sensory histidine kinase. Contains possible hydrophobic membrane spanning regions |
Promoters
| Comments:
|
| Unknown_sigma |
SigE promoter sequence is similar to whiG promoter. Paget MS et al. (1999) also showed that sigE is negatively autoretulated. |
| CseB |
sigE transcript was completely absent in cseB mutant; the sigE promoter is absolutely dependent on cseB. |
Terminator
GGGACCCGGAGAACCACGCAGGCGGCGAGTACGGCGGTGGCCGTGTTCGTCGCCCTCGGCGTTTCCCTGGCCG
>>>>>>>>>>>>> <<<<<<<<<<<<<<< |
3716178..3716212 |
41..75 |
- |
sigE |
Overview
|
