Operons:
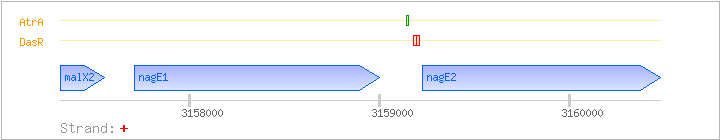
malX2-nagE1-nagE2
| malX2 |
SCO2905 |
SCE19A.05c, malX2 |
- |
3157315..3157548 |
hypothetical protein |
| nagE1 |
SCO2906 |
SCE19A.06, nagE1 |
+ |
3157707..3159002 |
putative PTS transmembrane component |
| nagE2 |
SCO2907 |
SCE19A.07, nagE2 |
+ |
3159229..3160479 |
putative PTS transmembrane component |
Promoters
| DasR |
- |
Negative |
-49:-34(from translation start site; upstream region of nagE2) |
3159180..3159195 |
ACAGGTCTACACCACT |
Rigali S, et al. (2004): HM, GS
Rigali S, et al. (2006): DB(by sqRT-PCR)
Nothaft H, et al. (2010): HM
|
| DasR |
- |
Negative |
-32:-14(from translation start site; upstream region of nagE2) |
3159197..3159212 |
AGTGGTGTAGACCACC |
Rigali S, et al. (2004): HM, GS
Rigali S, et al. (2006): DB(by sqRT-PCR)
Nothaft H, et al. (2010): HM
|
| AtrA |
- |
Positive |
-88:-73(from translation start site) |
3159141..3159155 |
GGAATCACGGGTTCC |
Nothaft H, et al. (2010): HM, GS, DB(by sqRT-PCR)
|
| Comments:
|
| DasR |
To build the matrices specific for PTSNag, we chose the cis-elements upstream crr-ptsI, between malX2 and nagE1, upstream nagE2, and upstream ptsH as training set of sequences. |
| DasR |
To build the matrices specific for PTSNag, we chose the cis-elements upstream crr-ptsI, between malX2 and nagE1, upstream nagE2, and upstream ptsH as training set of sequences. |
| AtrA |
AtrA binds to the predicted AtrA binding site upstream of nagE2 and that nagE2 transcript levels are strongly reduced in an atrA knock-out strain, demonstrating that AtrA trans-activates nagE2 transcription. |
Terminator
No Data
Overview
|
